Abstract
RNA-binding proteins (RBPs) play a critical role in regulating RNA splicing, polyadenylation, and translation. RBPs typically contain discrete RNA-binding domains (RBDs) that bind to specific nucleotide motifs. Given their important role in regulating gene expression, RBPs are often mutated or aberrantly expressed in a variety of malignancies, particularly hematological cancers. As a result, RBPs appear to be excellent therapeutic targets. However, given that most RBPs lack enzymatic activity, it has been challenging to develop targeted therapies.
Over the past several years we and others have used genetic and molecular biology approaches to understand the consequence of aberrant expression of the poly-C RBP, heterogeneous ribonucleoprotein K (hnRNP K), and determine the mechanisms by which it drives hematologic malignancies (Cancer Cell, 2015; NEJM, 2013, JNCI, 2020; and BioRxiv, 2021).
To develop a pharmacologic strategy to target hnRNP K's RNA binding activities, we developed a robust fluorescent anisotropy (FA) assay to evaluate hnRNP K's ability to bind target transcripts that are critical for tumorigenesis; such as, BRD4, MYC, RUNX1, and 7SK. We next performed small molecule screens using ~80,000 compounds and identified 34 compounds that inhibited hnRNP K's ability to bind RNA targets. To gain a deeper understanding of whether these compounds directly interact with hnRNP K to block its RNA binding functions, we performed SYPRO-orange thermal shift assays. Here we identified compounds that bound hnRNP K directly and altered hnRNP K thermostability. To evaluate association rate, dissociation rate, and KD constants, we next performed SPR assays. These experiments identified compounds bound hnRNP K with affinities between (350nM and 20uM).
We next performed cell-based studies, such as CETSA. Here we identified compounds that crossed the plasma membrane, directly engaged hnRNP K in vivo, and significantly stabilized hnRNP K. Molecular analyses revealed that cells treated with these lead compounds have reduced c-Myc expression, a direct mRNA target of hnRNP K. To develop a granular understanding of how these hnRNP K-small molecule interactions impact growth and cell survival, we performed high-throughput cell-based assays. These studies demonstrated that cells with inducible hnRNP K overexpression were sensitive to these small molecules.
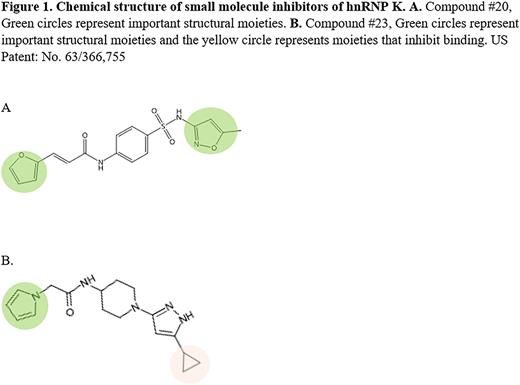
To begin medicinal chemistry approaches and to understand each compound's chemical requirements for binding to hnRNP K, we scanned molecules for reactive moieties. Here, we identified two compounds possessing amide groups within two carbons of either a furan or pyrrole moiety. These compounds, #20 and #23, respectively, were then chemically modified and FA assays were repeated in the presence of target mRNAs. Here, we observed that both the furan and isoxazole ring of compound #20 and pyrrole rings of compound #23 are critical for hnRNP K interactions, while the cyclopropane moiety on compound #23 is dispensable. Further analyses revealed additional structural analogs with either increased or decreased affinities and these results have been critical to our understanding of the importance of each moiety. Currently, we are using cryo-EM to evaluate positional interactions between hnRNP K (hnRNP K's RBDs) and these small molecules. These results will be used to further our medicinal chemistry approaches to generate more potent analogs.
Collectively, our results demonstrate that hnRNP K functions can be directly inhibited by small molecules that block its RNA binding functions and demonstrates that the development of effective therapies for RBPs is possible.
1. hnRNP K Is a Haploinsufficient Tumor Suppressor that Regulates Proliferation and Differentiation Programs in Hematologic Malignancies. Cancer Cell, 2015. 28(4): p. 486-499.
2. Cancer Genome Atlas Research Network, Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013 368(22):2059-74.
3. Uncovering the Role of RNA-Binding Protein hnRNP K in B-Cell Lymphomas. JNCI: Journal of the National Cancer Institute, 2020.
4. Heterogeneous nuclear ribonucleoprotein K is overexpressed in acute myeloid leukemia and causes myeloproliferative disease in mice via altered Runx1 splicing. BioRxiv. doi: https://doi.org/10.11.02.05.429385, 2021
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal